News
2025
November
-
Congratulations to Jiawei Hu and Wenxin Yang on joining our group as new graduate students! We are excited to have you on board and look forward to working with you.
-
Congratulations to Jiarong Li for winning Third Prize in the Oral Presentation and to Heqi Wang for achieving First Prize in the Poster Award at the Shanghai Society for Bioinformatics!

July
-
Welcome Feifei Sun, Wenxin Yang and Jiawei Hu for summer internship!
-
A research team led by Prof. Hong Li from the Shanghai Institute of Nutrition and Health (SINH), Chinese Academy of Sciences, has published a new study in Cell Reports Medicine presenting a novel computational framework for identifying compounds capable of overcoming resistance to pancancer immunotherapy.
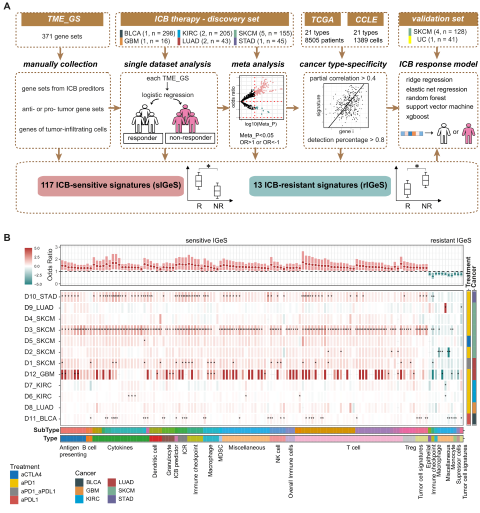
Immunotherapy has revolutionized cancer treatment, but many patients experience resistance. To tackle this issue, Fangyoumin Feng—first author of the study—and colleagues developed a method named IGeS-BS to prioritize candidate compounds that could enhance immunotherapeutic responses. Through meta-analysis of approximately 1,000 transcriptomes from immunotherapy-treated patients, the team identified 33 tumor microenvironment (TME) signatures capable of accurately predicting patient responses.
This study not only offers a powerful computational tool for drug discovery but also provides new therapeutic opportunities to overcome immune resistance in cancer. Prof. Hong Li, corresponding author, emphasized the translational potential of these findings in guiding combination immunotherapy strategies. The work was supported by the National Natural Science Foundation of China, the Natural Science Foundation of Shanghai, the CAS Youth Innovation Promotion Association, and the Shanghai Sailing Program.
June
-
Congratulations to Dr. Siyu Jing on continuing her impressive academic journey as a postdoctoral researcher in our lab!
-
Congratulations to Dr. Bihan Shen and Dr. Siyu Jing on their successful graduation! We are incredibly proud of your achievements and grateful for your contributions to the group. Wishing you all the best in your future endeavors!


March
- Cancer Systems Biology group contributes a research paper entitled “Quantifying and interpreting biologically meaningful spatial signatures within tumor microenvironments” in npj Precision Oncology.
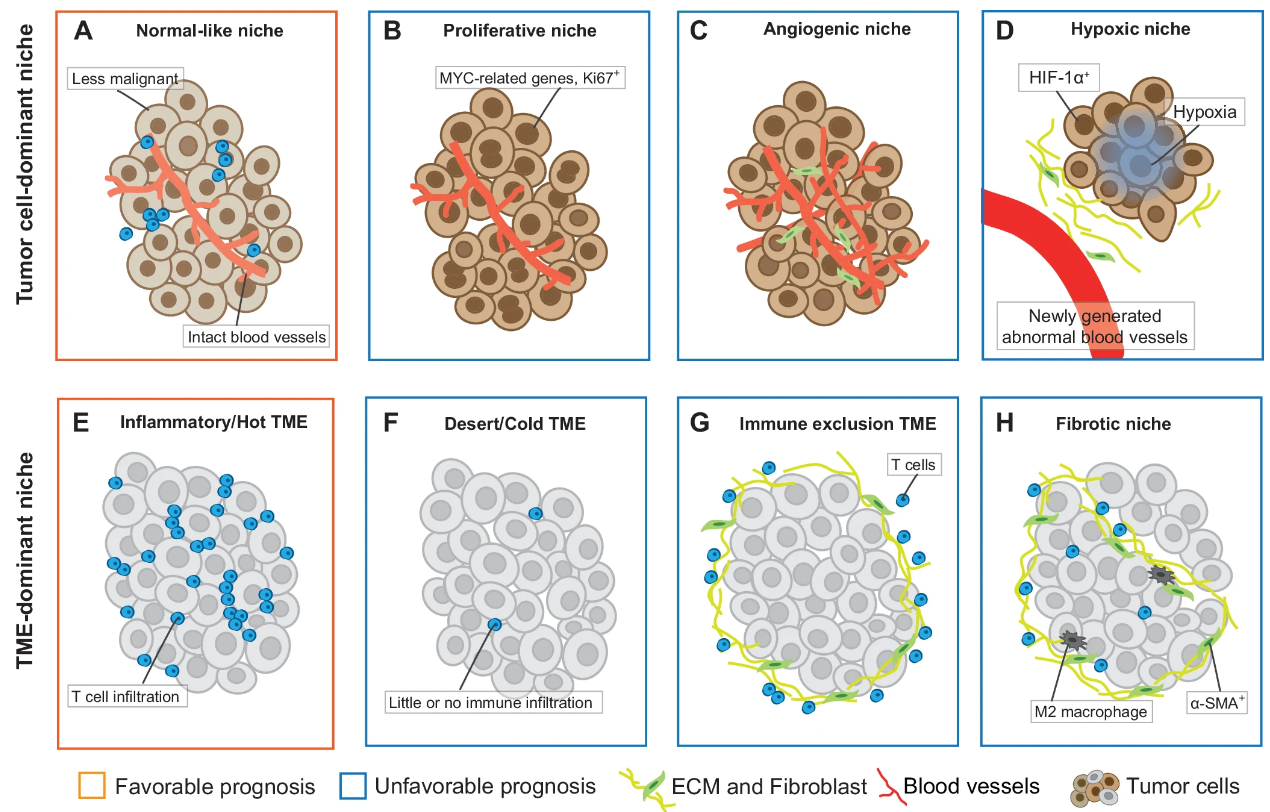
A research team led by Prof. Hong Li from Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences, publicated a new research paper on npj Precision Oncology. The tumor microenvironment (TME) plays a crucial role in orchestrating tumor cell behavior and cancer progression. Recent advances in spatial profiling technologies have uncovered novel spatial signatures, including univariate distribution patterns, bivariate spatial relationships, and higher-order structures. These signatures have the potential to revolutionize tumor mechanism and treatment. This review summarize the current state of spatial signature research, highlighting computational methods to uncover spatially relevant biological significance. We discuss the impact of these advances on fundamental cancer biology and translational research, address current challenges and future research directions. Si-yu Jing serve as the first authors, while Hong Li, both from the Cancer Systems Biology group at the Shanghai Institute of Nutrition and Health (SINH), CAS, serves as the corresponding author. This work was supported by the funding from National Natural Science Foundation of China, Natural Science Foundation of Shanghai and CAS Youth Innovation Promotion Association.
- Cancer Systems Biology group contributes a research paper entitled “SOAPy: a Python package to dissect spatial architecture, dynamics, and communication” in Genome Biology.
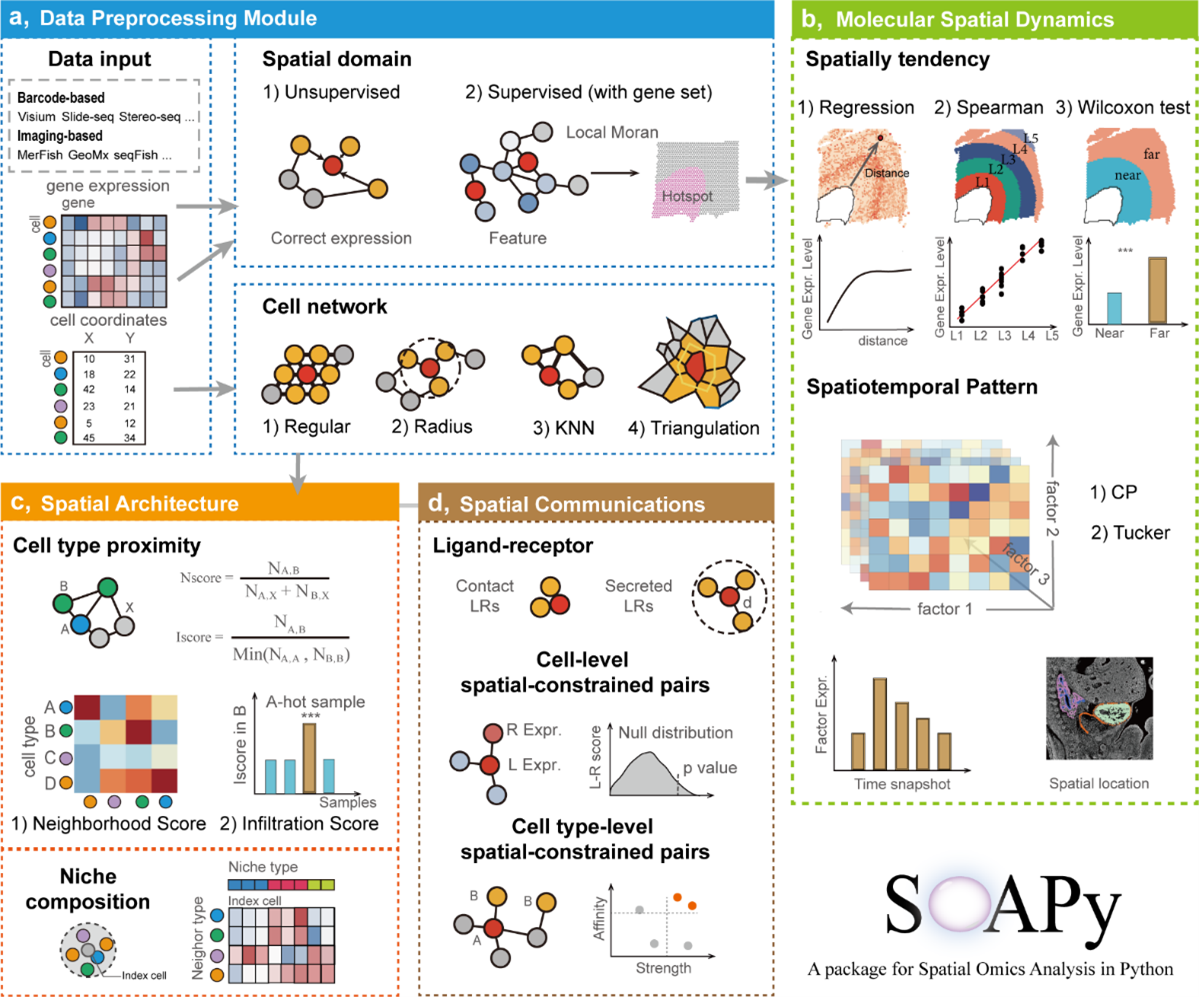
A research team led by Prof. Hong Li from Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences, developed and released a new toolkit named “SOAPy” for analyzing spatial omics data. Spatial organization and microenvironment are important for understanding the tissues in normal and disease states. Spatial omics especially spatial transcriptomes have become popular in recent years, but extracting useful information from these complex spatial data is still challenging. Previous algorithms were often designed for one specific task, so a toolkit for the integrative analysis of microenvironmental spatial organization is in an urgent need. Prof Li’s group developed a Python package named “SOAPy”. It provides methods for spatial domain identification, spatial expression tendency, spatiotemporal expression pattern, cellular co-localization, multi-cellular niches, and cell–cell communication. SOAPy can be applied to various spatial omics technologies and multiple areas in physiological and pathological contexts, such as tumor biology and development biology. In summary, this study presents a valuable tool for spatial omics analysis, facilitating the dissection of microenvironment architecture and deepening biological insights.
January
- Congratulations to Yan Zang, Yiling Qiu, and Yongqi Bei on joining our group as new graduate students! We are excited to have you on board and look forward to working with you.
2024
November
- Cancer Systems Biology group contributes a research paper entitled “A disentangled generative model for improved drug response prediction in patients via sample synthesis” in Journal of Pharmaceutical Analysis.
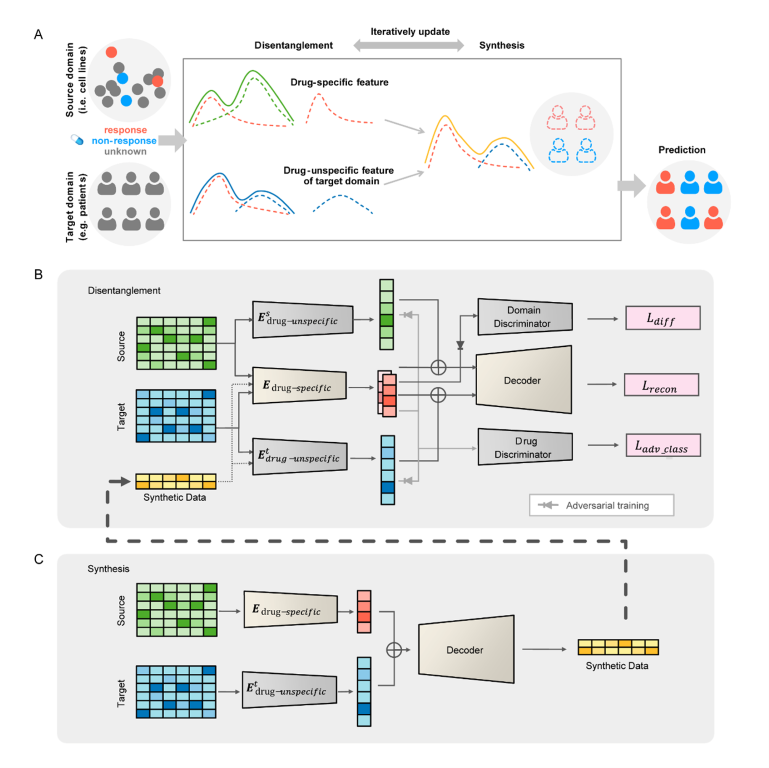
Personalized drug response prediction from molecular data is an important challenge in precision medicine for treating cancer. Computational methods have been widely explored and have become increasingly accurate in recent years. However, the clinical application of prediction methods is still in its infancy due to large discrepancies between preclinial models and patients. We present a novel disentangled synthesis transfer network (DiSyn) for drug response prediction specifically designed for transfer learning from preclinical models to clinical patients. DiSyn uses a domain separation network to disentangle drug response-related features, employs data synthesis technology to increase the sample size and iteratively trains for better feature disentanglement. DiSyn is pretrained on large-scale unlabeled cancer samples and validated by three datasets, TCGA, I-SPY2 and NIBR PDXE, achieving competitive accuracy with the state-of-the-art methods on cancer patients and mice. Furthermore, the application of DiSyn to thousands of breast cancer patients show the heterogeneity in drug responses and demonstrate its potential value in biomarker discovery and drug combination prediction.
Kunshi Li and Bihan Shen serve as the first authors, while Hong Li, both from the Cancer Systems Biology group at the Shanghai Institute of Nutrition and Health (SINH), CAS, serves as the corresponding author. This work was supported by the funding from National Natural Science Foundation of China, Natural Science Foundation of Shanghai and CAS Youth Innovation Promotion Association.
October
- Cancer Systems Biology group contributes a research paper entitled “Dual-view jointly learning improves personalized drug synergy prediction” in Bioinformatics.
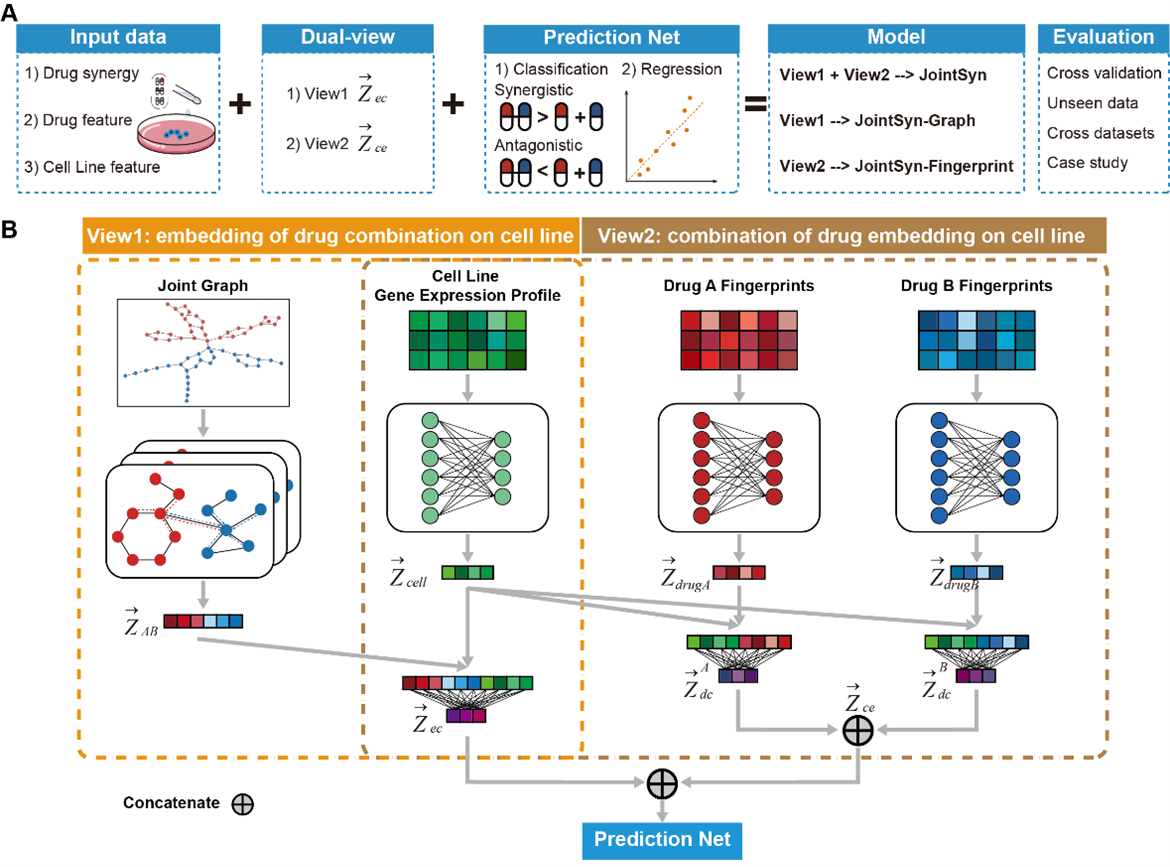
Accurate and robust estimation of the synergistic drug combination is important for medicine precision. Although some computational methods have been developed, some predictions are still unreliable especially for the cross-dataset predictions, due to the complex mechanism of drug combinations and heterogeneity of cancer samples. We have proposed JointSyn that utilizes dual-view jointly learning to predict sample-specific effects of drug combination from drug and cell features. JointSyn outperforms existing state-of-the-art methods in predictive accuracy and robustness across various benchmarks. Each view of JointSyn captures drug synergy-related characteristics and make complementary contributes to the final pre-diction of drug combination. Moreover, JointSyn with fine-tuning improves its generalization ability to predict a novel drug combination or cancer sample using a small number of experimental measure-ments. We also used JointSyn to generate an estimated atlas of drug synergy for pan-cancer and explored the differential pattern among cancers. These results demonstrate the potential of JointSyn to predict drug synergy, supporting the development of personalized combinatorial therapies.
Xueliang Li serves as the first author, while Hong Li, both from the Cancer Systems Biology group at the Shanghai Institute of Nutrition and Health (SINH), CAS, serves as the corresponding author. This work was supported by the funding from National Natural Science Foundation of China, Natural Science Foundation of Shanghai and CAS Youth Innovation Promotion Association.
August
- Interview with professor Li.
- Cancer Systems Biology group contributes a research paper entitled “Spatial multiomics reveals a subpopulation of fibroblasts associated with cancer stemness in human hepatocellular carcinoma” in Genome Medicine.
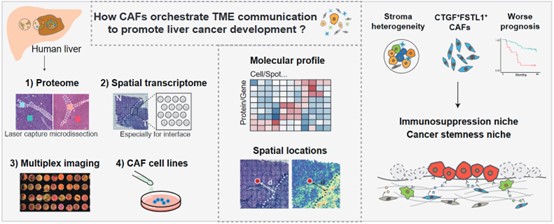
Cancer-associated fibroblasts (CAFs) are the prominent cell type in the tumor microenvironment (TME), and CAF subsets have been identified in various tumors. However, how CAFs spatially coordinate other cell populations within the liver TME to promote cancer progression remain unclear. In the study, researchers combined multi-region proteomics (6 patients, 24 samples), 10X Genomics Visium spatial transcriptomics (11 patients, 25 samples) and multiplexed imaging (92 patients, 264 samples) technologies to decipher the expression heterogeneity, functional diversity, spatial distribution, colocalization and interaction of fibroblasts. The newly identified CAF subpopulation was validated by cells isolated from 5 liver cancer patients and in vitro functional assays. The study identified a liver CAF subpopulation, marked by the expression of COL1A2, COL4A1, COL4A2, CTGF and FSTL1 and named F5-CAF. F5-CAF preferentially located within and around tumor nests and colocalizes with cancer cells with higher stemness in hepatocellular carcinoma (HCC). Multiplexed staining of 92 patients and the bulk transcriptome of 371 patients demonstrated that the abundance of F5-CAFs in HCC was associated with a worse prognosis. Further in vitro experiments showed that F5-CAFs isolated from liver cancer patients can promote the proliferation and stemness of HCC cells.
Siyu Jing from Cancer Systems Biology group at Shanghai Institute of Nutrition and Health (SINH), CAS, Dan Liu, Na Feng and Hui Dong from National Center for Liver Cancer, Eastern Hepatobiliary Surgery Hospital serves as the co-corresponding authors. This work was supported by the funding from Chinese Academy of Sciences, the National Key Research and Development Project, the National Natural Science Foundation of China, CAS Youth Innovation Promotion Association, Shanghai Science and Technology Committee, Shanghai Municipal Science and Technology Major Project, and the Shanghai Sailing Program.
For more information, please refer to: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-024-01367-8
July
March
- Congrats to professor Li for being awarded for the 2024 Shanghai Science and Technology System “March 8th Red-Banner Pacesetter” !
- Cancer Systems Biology group contributes a research paper entitled “Applying machine learning for multi-individual Raman spectroscopic data to identify different stages of proliferating human hepatocytes” in iScience.
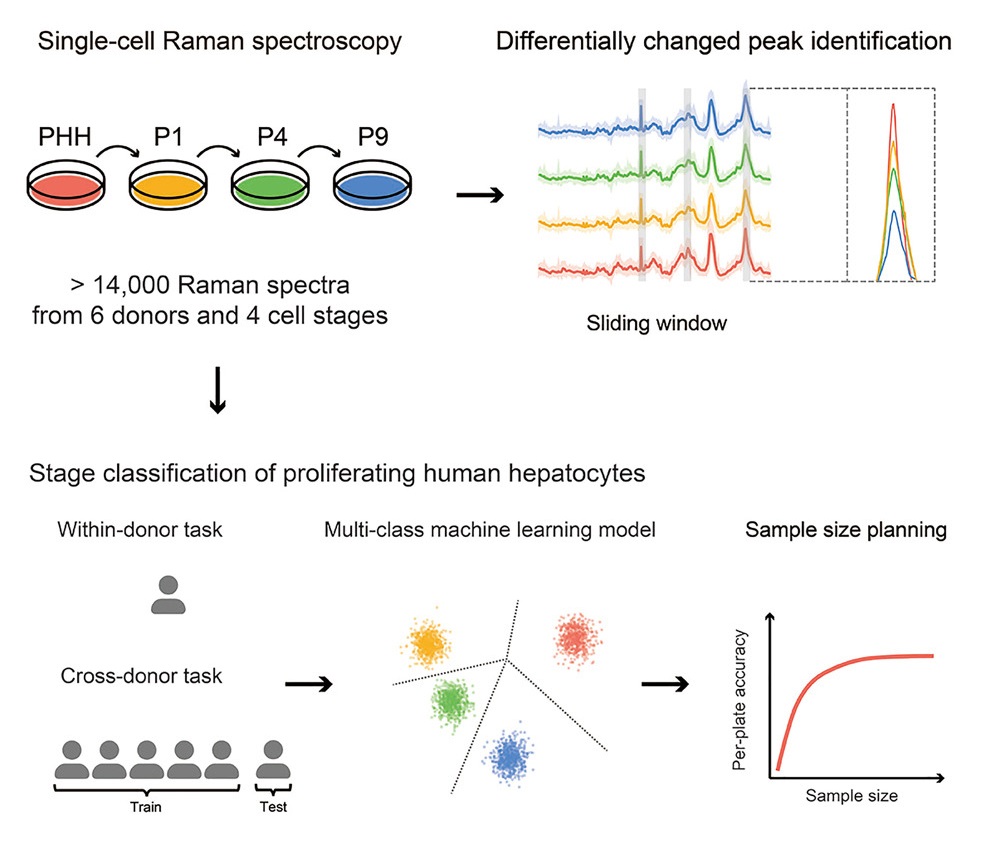
Cell therapy using proliferating human hepatocytes (ProliHHs) is an effective treatment approach for advanced liver diseases. However, rapid and accurate identification of high-quality ProliHHs from different donors is challenging due to individual heterogeneity. Here, we developed a machine learning framework to integrate single-cell Raman spectroscopy from multiple donors and identify different stages of ProliHHs. A repository of more than 14,000 Raman spectra, consisting of primary human hepatocytes (PHHs) and different passages of ProliHHs from six donors, was generated. Using a sliding window algorithm, potential biomarkers distinguishing the different cell stages were identified through differential analysis. Leveraging machine learning models, accurate classification of cell stages was achieved in both within-donor and cross-donor prediction tasks. Furthermore, the study assessed the relationship between donor and cell numbers and its impact on prediction accuracy, facilitating improved quality control design. A similar workflow can also be extended to encompass other cell types.
January
2023
December
- We welcome new graduate students Jingxuan Cai and Jiao Yuan to join our group!
- Congrats to Bihan Shen for being awarded for “2023 Best Poster Award” in GRAD !
November
- Welcome Jingxuan Cai and Jiao Yuan for Lab Rotation!
- Welcome Yongqi Bei, Huicheng Ye and Feifei Sun for internship!
October
- Congrats to the success of the 2023 Bioinformatics and Computational Biology Young Talents Frontier Forum!
- Congrats to professor Li for receiving the 2023 Young Star Award of Shanghai Society for Bioinformatics.
September
- Welcome Qianxi Jia and Xintong Huang for Lab Rotation!
July
- Welcome Fei Guo, Wenwen Zhang and Chenyihang Xiong for summer internship!
- Welcome laboratory manager Zhixuan Tang to join our group!
- Congrats to Bihan Shen and Siyu Jing for being awarded for the 2023 Shanghai Branch of the Chinese Academy of Sciences “Rising Star in Technological Breakthroughs Award” !
- Congrats to professor Li for receiving the Second Outstanding Scientific Research Achievement Award for Female Scientists of the Chinese Biophysical Society of China.
May
- Cancer Systems Biology group contributes a review entitled “Single-cell and spatially resolved transcriptomics for liver biology” in Hepatology.
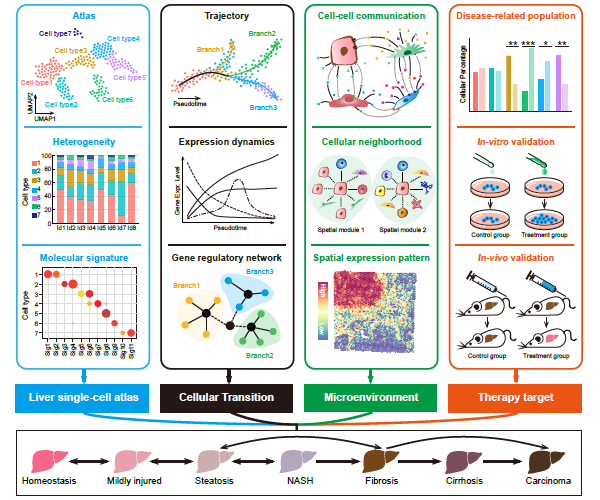
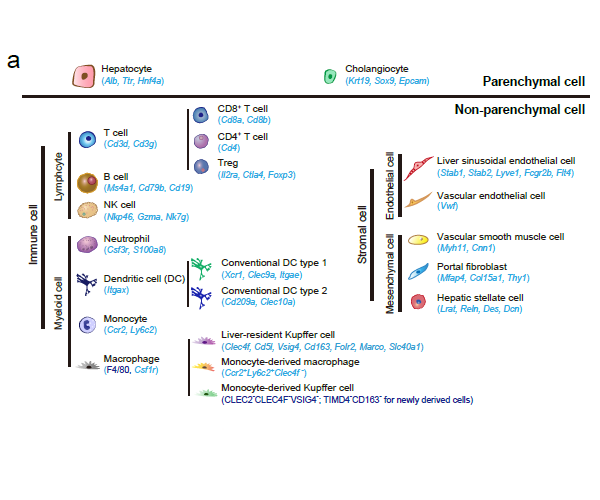
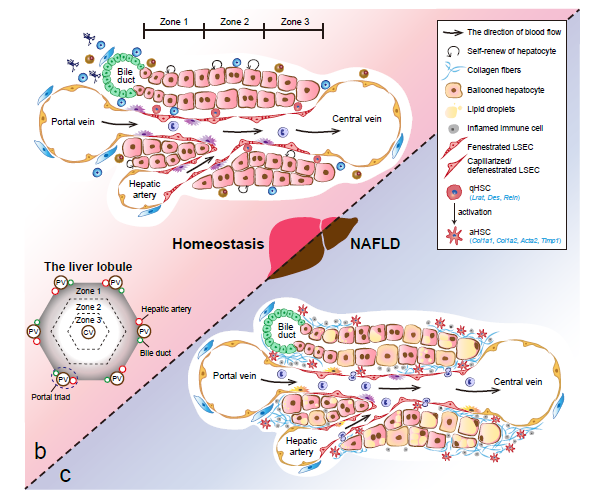
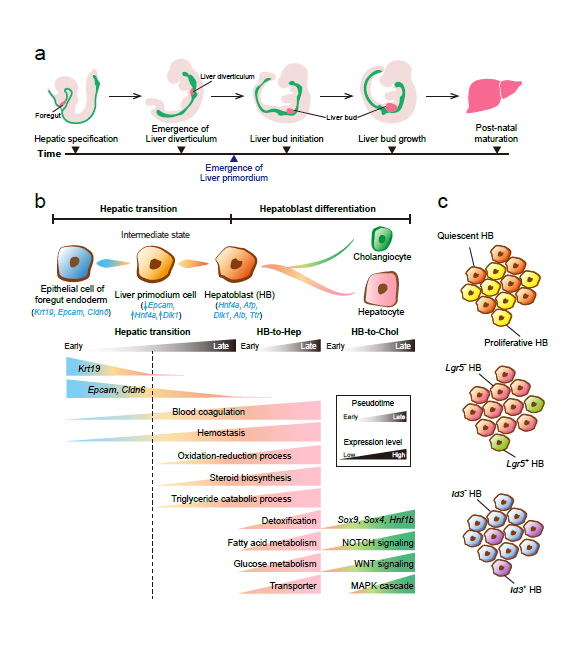
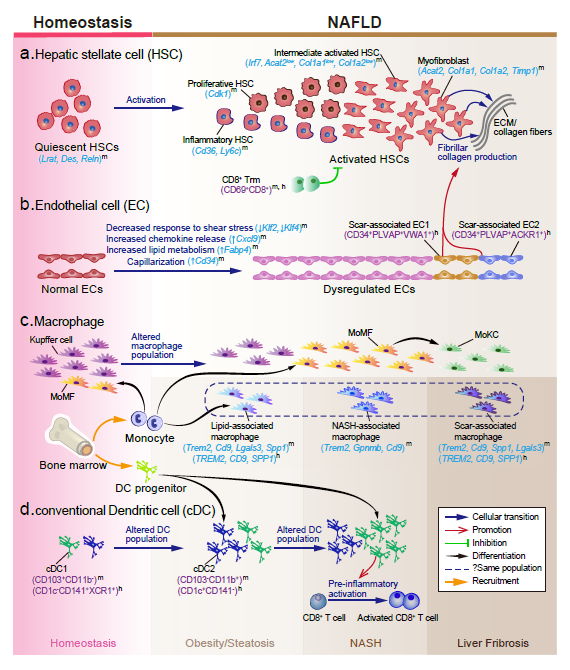
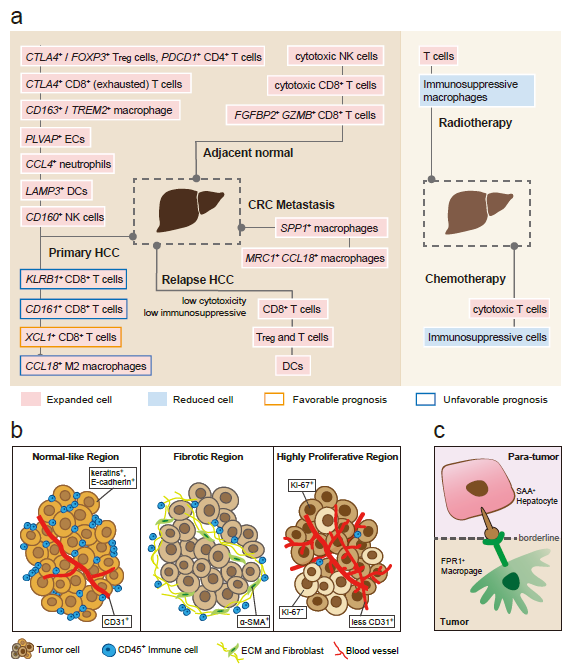
Recently, our lab contributes a review article entitled “Single-cell and spatially resolved transcriptomics for liver biology”. We introduced the basic knowledge about single-cell and spatially resolved techniques and the state-of-art computational tools used for data processing. Four common scenes were summarized when applying these techniques in the study of liver biology: (a) constructing the liver single-cell atlas; (b) the deduction of cellular transitions; (c) the characterization of tissue microenvironments; (d) the identification of potential therapy targets (Fig. 1). The four scenarios were seen in the settings of liver homeostasis, embryonic liver development, liver regeneration, non-alcoholic liver disease (NAFLD) and liver cancer. In each topic, we presented the cutting-edge progresses uncovered by single-cell and/or spatial techniques, such as the single-cell atlas of the liver under both diseased and homeostatic conditions (Fig. 2a), transcriptomic zonal patterns of hepatic cells (Fig. 2b), developmental trajectory of embryonic liver (Fig. 3), cell identity conversion of parenchymal cells upon injury, NAFLD-associated cell subtypes (Fig. 4), and cancer-associated cell populations within tumor microenvironments (Fig. 5). We highlighted the computational strategies that help achieve these accomplishments; and discussed the outstanding questions remaining elusive. Lastly, we pointed out the challenges when applying these technologies to the liver biology, and for each of which, we proposed possible solutions.
- Welcome Jingxuan Cai for summer internship!
April
- Congrats to professor Li for receiving the 2023 China Cell Biology Society-TianNeng Life Science Young Female Scientist Career Development Fund, category B !
February
- Cancer Systems Biology group contributes a research paper entitled “Kupffer cell-derived IL-6 is repurposed for hepatocyte dedifferentiation via activating progenitor genes from injury-specific enhancers” in Cell Stem Cell.
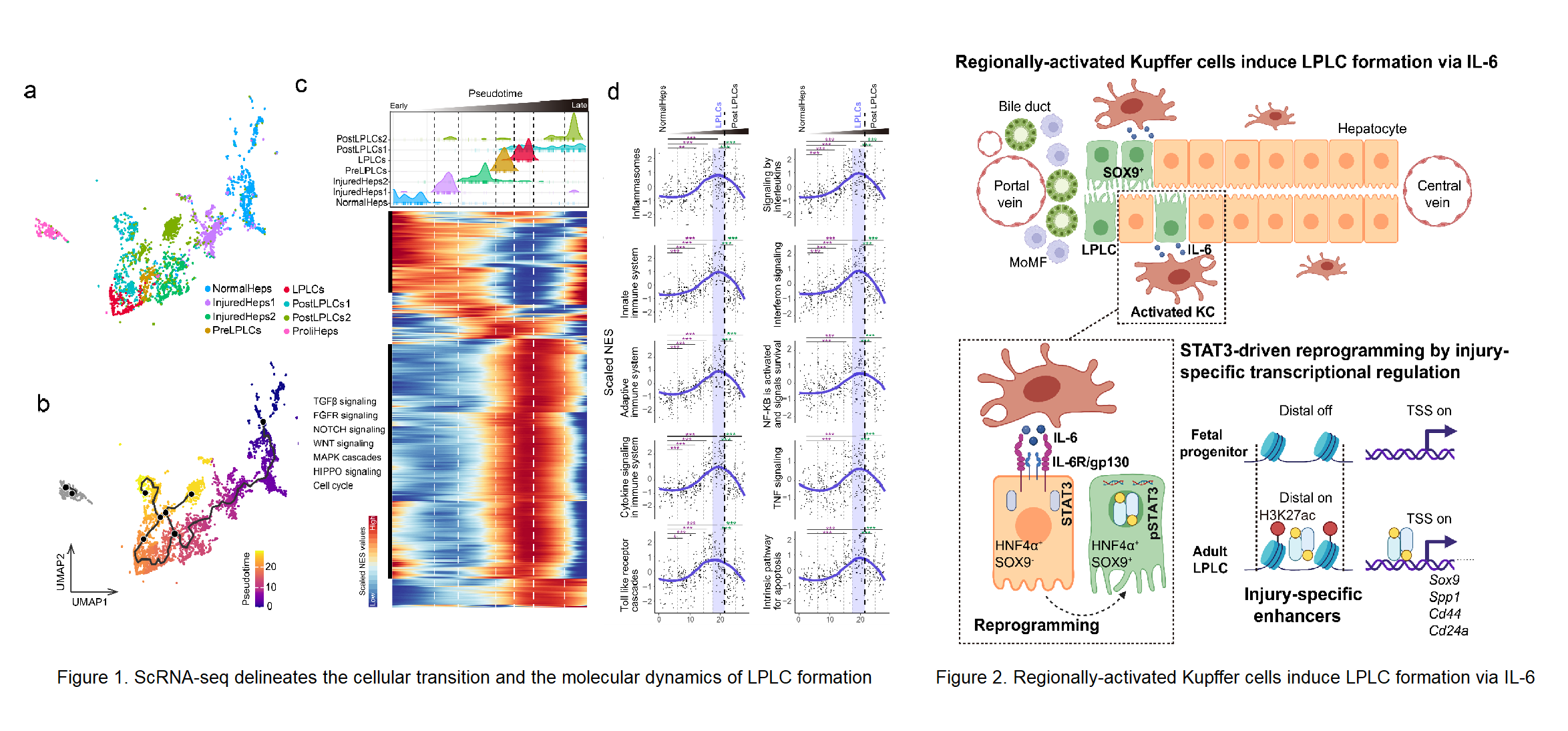
Stem cell-independent reprogramming of differentiated cells has recently been identified as an important paradigm for repairing injured tissues. Following periportal injury, mature hepatocytes re-activate reprogramming/progenitor-related genes (RRGs) and dedifferentiate into liver progenitor-like cells (LPLCs) in both mice and humans, which contribute remarkably to regeneration. However, it remains unknown which and how external factors trigger hepatocyte reprogramming. Here, by employing single-cell transcriptional profiling we reconstructed the cellular trajectory of LPLC formation and deciphered the key pathways underlying cellular state transition during the process (Figure 1). We found that immune related pathways were highly correlated with LPLC formation. By utilizing lineage-specific deletion tools, we uncovered that periportal-specific LPLC formation was initiated by regionally activated Kupffer cells but not peripheral monocyte-derived macrophages (Figure 2). Unexpectedly, using in vivo screening, the proinflammatory factor IL-6 was identified as the niche signal repurposed for RRG induction via STAT3 activation, which drove RRG expression through binding to their pre-accessible enhancers. Notably, RRGs were activated through injury-specific rather than liver embryogenesis-related enhancers. Collectively, these findings depict an injury-specific niche signal and the inflammation-mediated transcription in driving the conversion of hepatocytes into a progenitor phenotype (Figure 2). Lu Li, Lei Cui from Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Center for Excellence in Molecular Cell Science, CAS and Ping Lin from Cancer Systems Biology group at Shanghai Institute of Nutrition and Health (SINH), CAS serve as co-first authors. Professor Lijian Hui from SIBCB, Center for Excellence in Molecular Cell Science, CAS, Professor Hong Li from Cancer Systems Biology group at SINH, CAS and Professor Yixue Li from SINH, CAS serves as the co-corresponding authors. This work was supported by the funding from Chinese Academy of Sciences, the National Key Research and Development Project, the National Natural Science Foundation of China, CAS Youth Innovation Promotion Association, Shanghai Science and Technology Committee, Shanghai Municipal Science and Technology Major Project, and the Shanghai Sailing Program.
For more information, please refer to: BioArt iNature
- Congrats to Siyu Jing for being awarded for “2022 Best Research Report Award” in GRAD !

2022
December
-
Congrats to professor Li for being awarded for “2022 Excellent Member Award, Youth Innovation Promotion Association, Chinese Academy of Sciences” !
-
Cancer Systems Biology group contributes a benchmarking paper entitled “A systematic assessment of deep learning methods for drug response prediction: from in vitro to clinical applications” in Briefings in Bioinformatics.
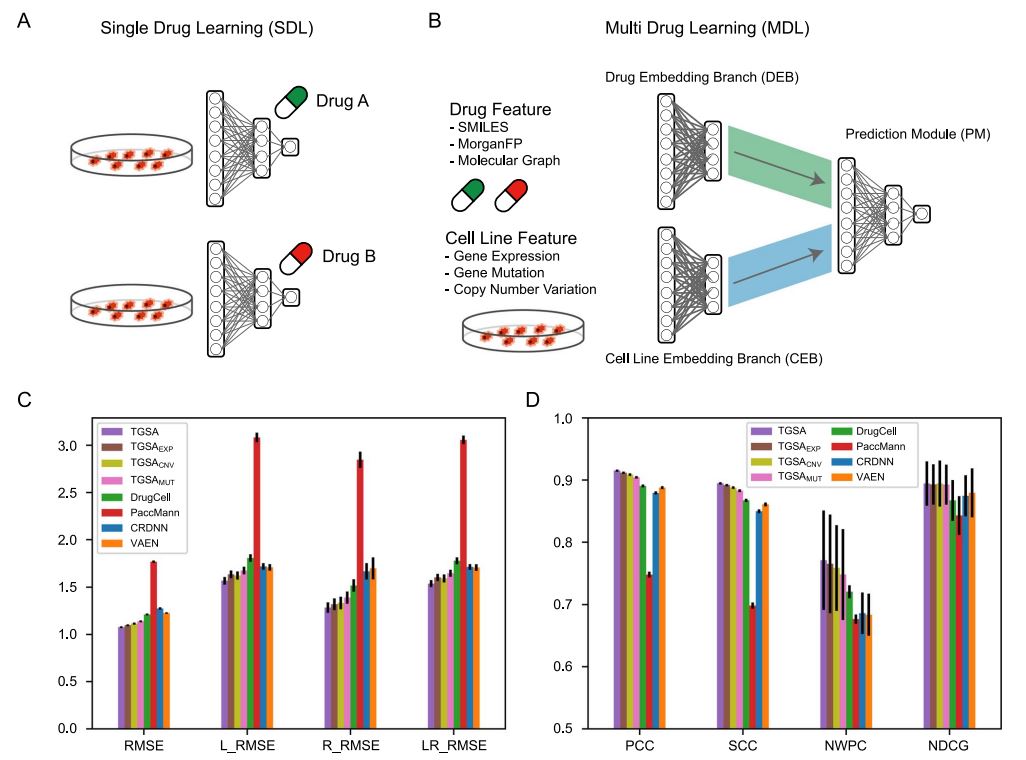
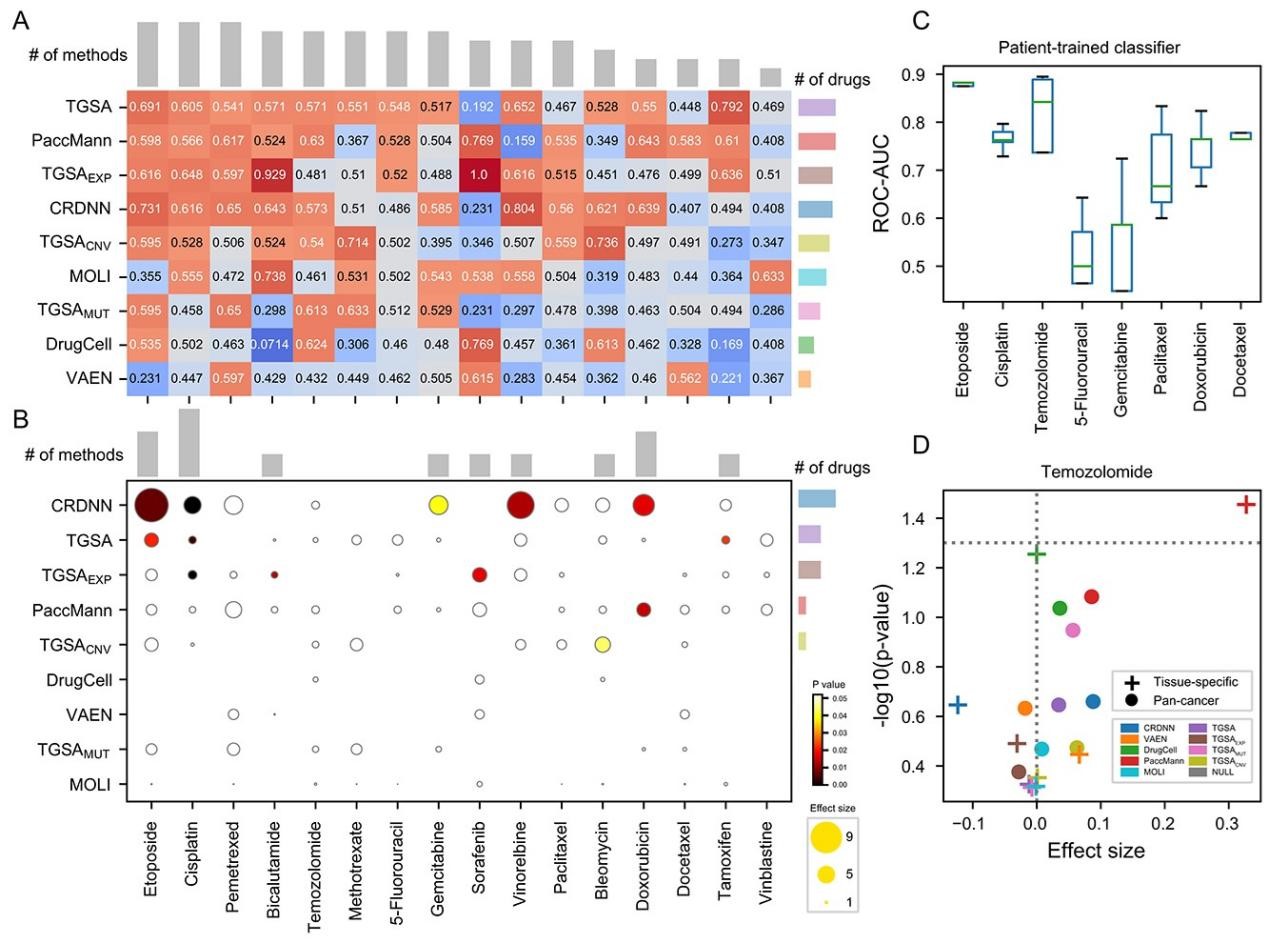
Drug sensitivity prediction is an important and challenging problem in both bioinformatics and translational medicine. Various categories of methods have been developed and compared. However, previous benchmark studies have rarely discussed the predictive ability of preclinical models on clinical patients, and not comprehensively covered the paradigms of deep learning methods. To provide a more rigorous assessment, the performance of six representative deep learning methods for drug response prediction in multiple application scenarios was benchmarked, including the overall prediction accuracyusing nine evaluation metrics, predictability of each drug, potential associated factors and transferability to clinical cohorts. Most methods show promising prediction within cell line datasets (Figure 1). TGSA, with its lower time cost and better performance, is recommended. The performance metrics decrease when applying models trained on cell lines to patients (Figure 2), due to the differences between cell lines and patients, the lack of drug sensitivity related features and the reduction of statistical power due to small sample sizes. However, clinical response on some drugs can still be distinguished using CRDNN and TGSA models. These assessments provide a guidance for researchers to choose appropriate methods, as well as insights into future directions for the development of more effective methods in clinical scenarios. Bihan Shen and Fangyoumin Feng serve as co-first authors. Professor Hong Li serves as the corresponding author. Kunshi Li and Ping Lin also contributed to this work. We thank Bio-Med Big Data Center at Shanghai Institute of Nutrition and Health, Xin Li (Shanghai Institute of Nutrition and Health, CAS) and Wenju Cai (Guizhou science data center Gui’an Supercomputing Center) for providing computational resource and discussions. This work is supported by National Natural Science Foundation of China, Natural Science Foundation of Shanghai, National Key R&D Program of China, CAS Youth Innovation Promotion Association and the development fund for Shanghai talents.
- We welcome new graduate students Wanhong Chen, Xufeng Chen and Xueliang Li to join our group!
November
- Welcome Xufeng Chen for Lab Rotation!
August
- Welcome Wanhong Chen, Xueliang Li, Zhaozhen Du for Lab Rotation!
July
- The Cancer Systems Biology (CSB) group is recruiting bioinformatics graduate students, postdocs, and research scientists. Please contact us by email (lihong01
 sinh.ac.cn).
sinh.ac.cn).
- Congrats to professor Li for being awarded for the 2021 Shanghai Science and Technology System “Youth May Fourth Medal” !
- Congrats to Dr. Feng for the completion of Postdoctoral Fellowships!